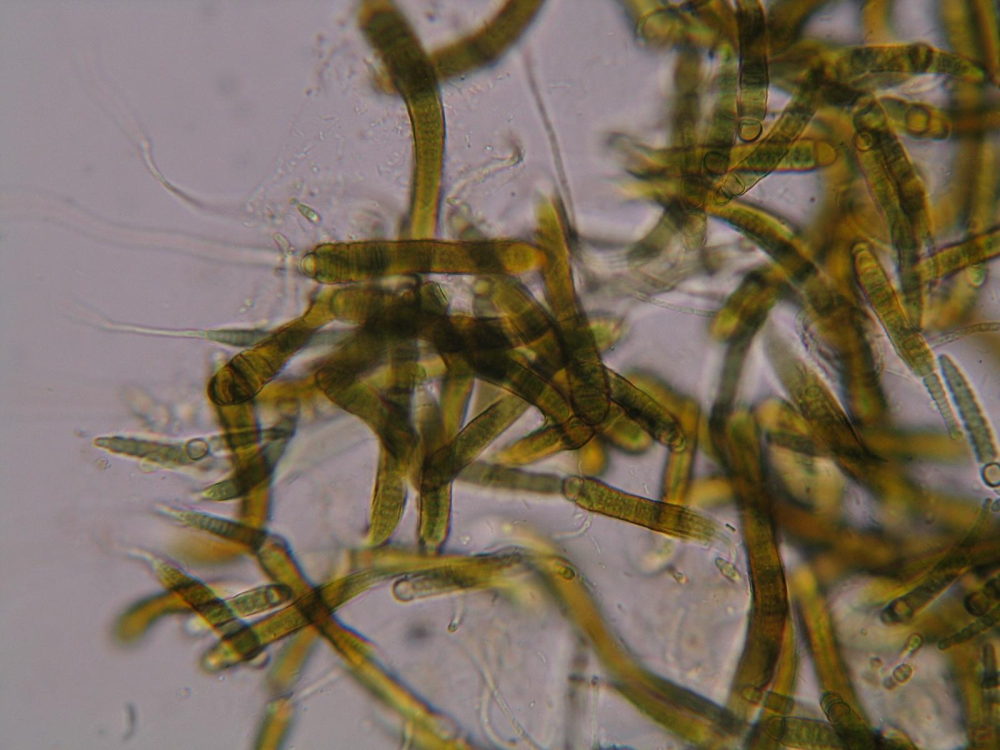
Microbial biodiversity is of fundamental importance to ecosystem functioning. To gain insights into the hidden world of microbes, culturing techniques still hold an important place in many research strategies, as they allow to perform experiments with particular organisms in isolation. For example, the use of cultures enables researchers to test substrates and waste products of strains, look at their enzyme expression rate in response to an environmental stimulus with mRNA sequencing, study interactions with other organisms, or infer their phylogenetic placement in the the tree of life using by comparing DNA sequences.
Especially Antarctic microbes have long interested scientists for their ability to thrive in the extreme cold, dry and saline environments that are common on the continent. The peculiar geographic location of Antarctica, isolated by the Southern Ocean and Circum Antarctic polar vortex winds, also created the conditions for the evolution of unique microbial diversity, while it significantly limited the presence (and thus impact) of humans. This is reflected in the relative high number of endemic taxa, with evidence indicating that at least some of these lineages trace their occupation of the frozen continent back to the early stages of the breakup of Gondwana.
However, obtaining living material from the last true wilderness on Earth is not straightforward, and most Antarctic microbes are slow growing, with very specific nutrient and culture requirements. Therefore, several dedicated culture collections exist, that maintain living specimen collected over the last two decades. This allows continuity in experiments (i.e. they can be repeated with the same strains), or facilitates comparison with new specimen in a research field that is in continuous flux driven by rapidly evolving technologies.
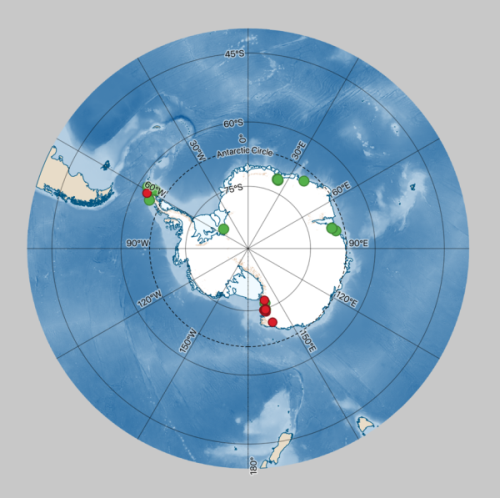
We used the framework of the microbial Antarctic Resource System (mARS) to create additional visibility for two culture collections, being the BCCM/ULC public Cyanobacteria Collection, supported by the Belgian Science Policy Office (Belspo) and hosted at the University of Liège, and the MNA-CIBAN Bacterial Collection of the Italian National Antarctic Museum.
In close collaboration with the hosting institutions, we used a standardized approach to compile a dataset with all the available information on each individual culture in the collection. This way, mARS helps to preserve data and connect researchers to the existing wealth of microbial resources that can be effectively browsed to identify any strain of interest. In addition, the data resources also serve as a reference for the occurrence of specific microbial species, which is relatively rare at a species or population level for prokaryotes.